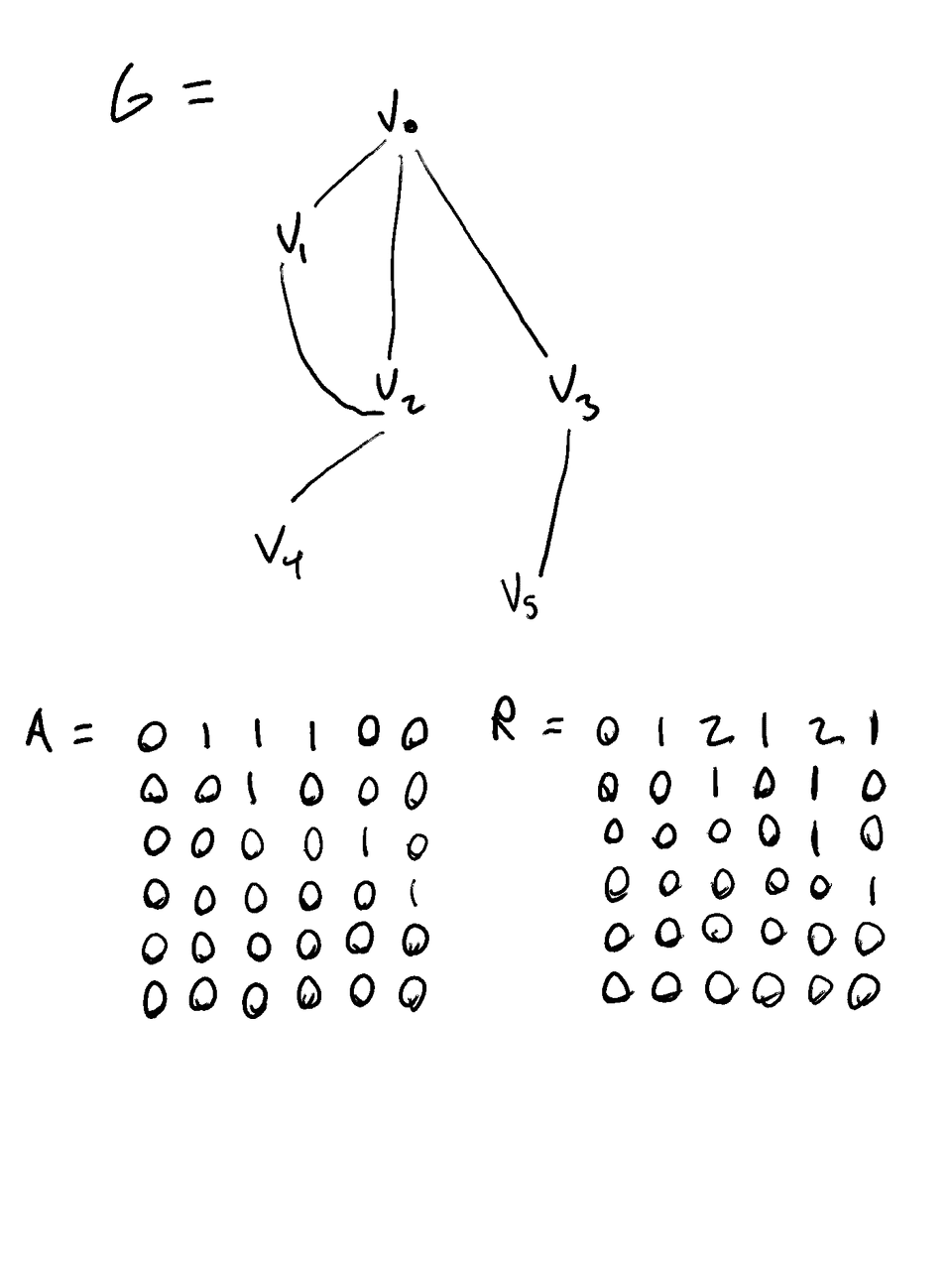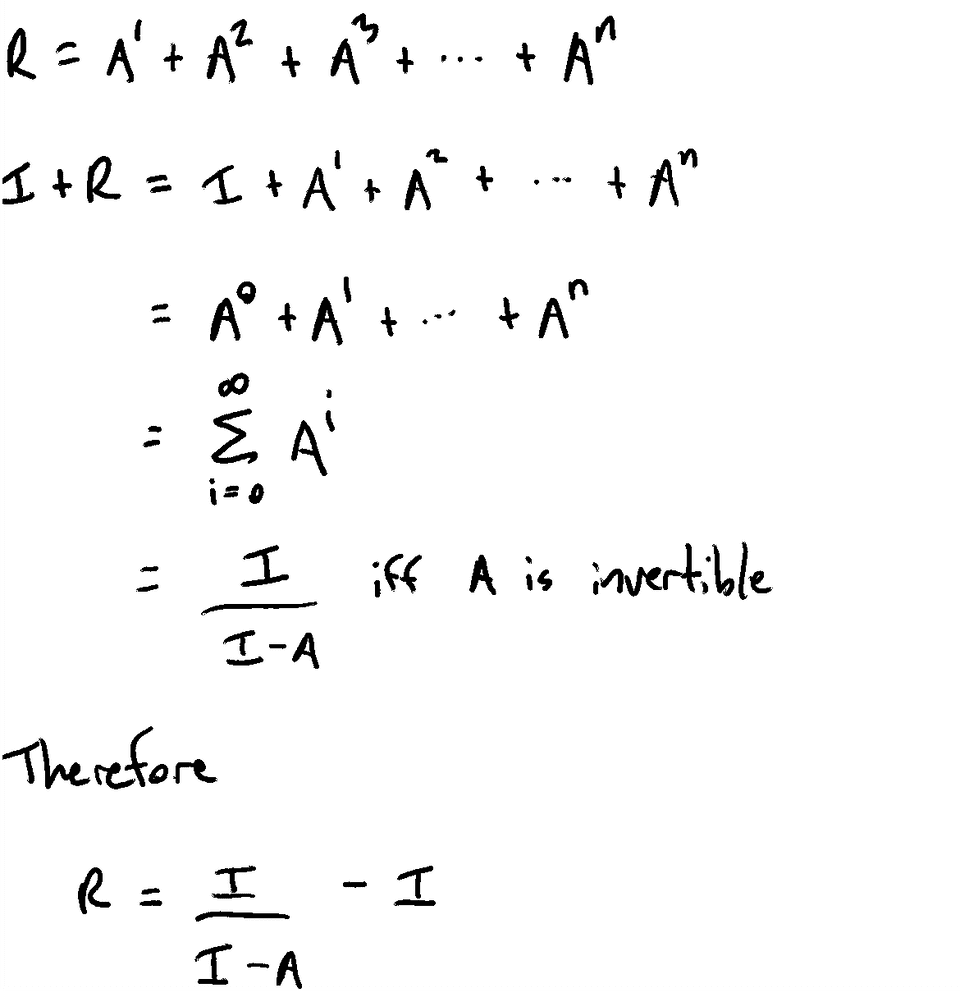We describe an algorithm for performing efficient reachability queries on DAGs using reachability matrices.
Constraints on our Graphs
The graphs supported by this algorithm have the following constraints:
- Directed
- Acyclic
- Single connected component
- Distinct edges
Definitions
- Let G be our graph
-
Let A be the Adjacency matrix of G
- Where A[i][j] == 1 iff Vi is a parent of Vj, 0 otherwise
-
Let A^n be the Walkability matrix
- Where A^n[i][j] == the number of walks of length n from Vi to Vj
-
Let R be the reachability matrix
- Where R[i][j] == the number of walks of any length from Vi to Vj
Example
Queries
getAncestors(Vertex v)
The column for a vertex encodes the information about which other vertices are ancestors.
Retrieve the column for v
For each vertex j
If col[j] >= 1, j is an ancestor of vgetDescendants(Vertex v)
The row for a vertex encodes the information about which other vertices are descendants.
Retrieve row for v
for each vertex j
If row[j] >= 1, j is a descendantdoesPathExist(Vertex to, Vertex from)
Return R[from][to] >= 1Constructing the Reachability Matrix
Proof the Series Converges
The series must converge because our graph does not allow cycles. Therefore, every walk must be finite and there must be some n where A^n is the empty matrix. Therefore, the series must converge.
Mutations
addEdge(Vertex a, Vertex b)
We want to add an edge from a to b.
Update a’s ancestors
We need to update a’s ancestors to show they have paths to b and all of b’s descendants. For every descendant of b, j, there is now an additional path from every ancestor to j.
The row for b contains the entries for every vertex b can reach. We can add the row for b to the row’s for a and every ancestor of a.
For each ancestor i of b:
row_i = row_i + row_bUpdate b’s descendants
We need to update b’s descendants to show there are paths from a and all of a’s ancestors. For every ancestor of a, i, there is now an additional path from i to every descendant.
The column for a contains the entries for every vertex a can be reached from. We can add the column for a to the columns for every descendant of b.
For each descendant j of b:
col_j = col_j + col_aUpdate R[a][b]
R[a][b] = R[a][b] + 1removeEdge(Vertex a, Vertex b)
Remove an edge from a to b.
Update a’s ancestors
We need to update a’s ancestors to remove paths to b and all of b’s descendants.
For each ancestor i of a:
row_i = row_i - row_bUpdate b’s descendants
We need to update b’s descendants to remove paths from a and all of a’s ancestors.
For each descendant j of b:
col_j = col_j - col_aUpdate R[a][b]
R[a][b] = R[a][b] - 1addVertex(Vertex v, List parents)
To add a vertex, we add an empty row and column to the matrix. If n is the number of vertices, R is an n x n matrix. When we add a vertex, R becomes an n+1 x n+1 matrix. Each parent must be distinct.
Add row and column
For each parent:
addEdge(parent, v)removeVertex(Vertex v)
To remove a vertex v, it must be a leaf. Otherwise, the graph would become disconnected.
Remove the row and column for vTime Complexity for Adding and Removing an Edge
Dimension of our Reachability Matrix
Let n be the number of vertices, then we have an n x n reachability matrix
Remove the leaf rows
Let k be the number of leaves
- Every leaf will always have a sparse row
- Remove k - 1 rows (at most we could be affecting 1 leaf with our mutation)
- We now have an n x (n - k + 1) matrix
How many entries in our matrix can be non-zero?
Observe that if we sort our vertex columns by ascending depth in the graph
-
We get an upper diagonal matrix
- Half of our entries will be 0
- There are at most n * (n - k + 1) / 2 entries that could be populated
Adding and removing an edge is an O(n * (n - k + 1) / 2) operation where n is the number of vertices and k is the number of leaves.
Note that each operation is a scalar addition or subtraction.